RNA secondary structure prediction based on SHAPE data in helix regions
Abstract
RNA molecules play important and fundamental roles in biological processes. Frequently, the functional form of single-stranded RNA molecules requires a specific tertiary structure. Classically, RNA structure determination has mostly been accomplished by X-Ray crystallography or Nuclear Magnetic Resonance approaches. These experimental methods are time consuming and expensive. In the past two decades, some computational methods and algorithms have been developed for RNA secondary structure prediction. In these algorithms, minimum free energy is known as the best criterion. However, the results of algorithms show that minimum free energy is not a sufficient criterion to predict RNA secondary structure. These algorithms need some additional knowledge about the structure, which has to be added in the methods. Recently, the information obtained from some experimental data, called SHAPE, can greatly improve the consistency between the native and predicted RNA secondary structure.
In this paper, we investigate the influence of SHAPE data on four types of RNA substructures, helices, and loops, base pairs from the start and end of helices and two base pairs from the start and end of helices. The results show that SHAPE data in helix regions can improve the prediction. We represent a new method to apply SHAPE data in helix regions for finding RNA secondary structure. Finally, we compare the results of the method on a set of RNAs to predict minimum free energy structure based on considering all SHAPE data and only SHAPE data in helix regions as pseudo free energy and without SHAPE data (without any pseudo free energy). The results show that RNA secondary structure prediction based on considering only SHAPE data in helix regions is more successful than not considering SHAPE data and it provides competitive results in comparison with considering all SHAPE data.
RNAStructure
1. Please download RNAstructure.
2. Set the DATAPATH as "path/to/RNAstructure/data_table/".
3.Please note that the requirment files (fold.exe, input file, itr-RNAstructure.pl) should be in tha same folder.
Input files:
1. The format of a sequence file is *.seq.
2. The native structure format is *.dbn and the algorithm needs this file to compute the accuracy of prediction.
3. The format of Shape Data is *.shape.
Output file:
1. The output file is sim-last.text.
RNAstructure predicts several structures. We select the first one becuase minimum free energy of it is lower than the others.
figure1: The input files.
figure2: The output files and the number of algorithm iterations.
GTfold
1. Please download GTfold software and Data Folder.
2. Please note that the requirment files (gtmfe, Data folder, itr-Gtfold.pl) should be in tha same folder.
Input files:
1. The format of a sequence file is *.seq.
2. The native structure format is *.dbn and the algorithm needs this file to compute the accuracy of prediction.
3. The format of shape data is *.shape.
Output file:
1. The output file is sim-last.text.
GTfold predicts only one structure.
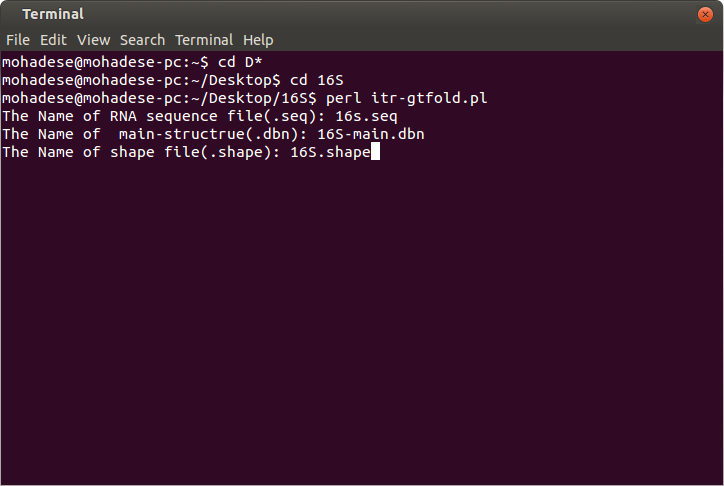
figure1: The input files.
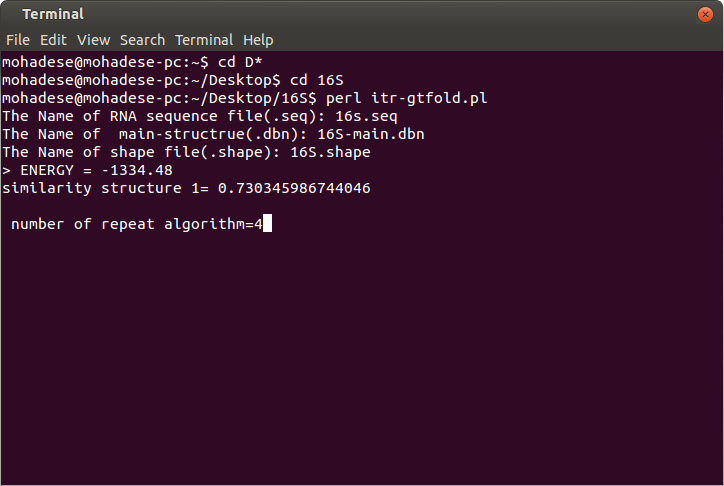
figure2: The output file and the number of algorithm iterations.