Introduction
The data set used in the paper contains the PPI networks of E. coli and S. cerevisiae which are extracted from the Database of
Interacting Proteins (DIP). To label the proteins as essential/non-essential, the essential genes data of these species are collected
from DEG database. Further description of the data is as follows.
PPI networks
In the raw data of protein-protein interactions in DIP [1] for E. coli, 12246 interactions between 2924 proteins have
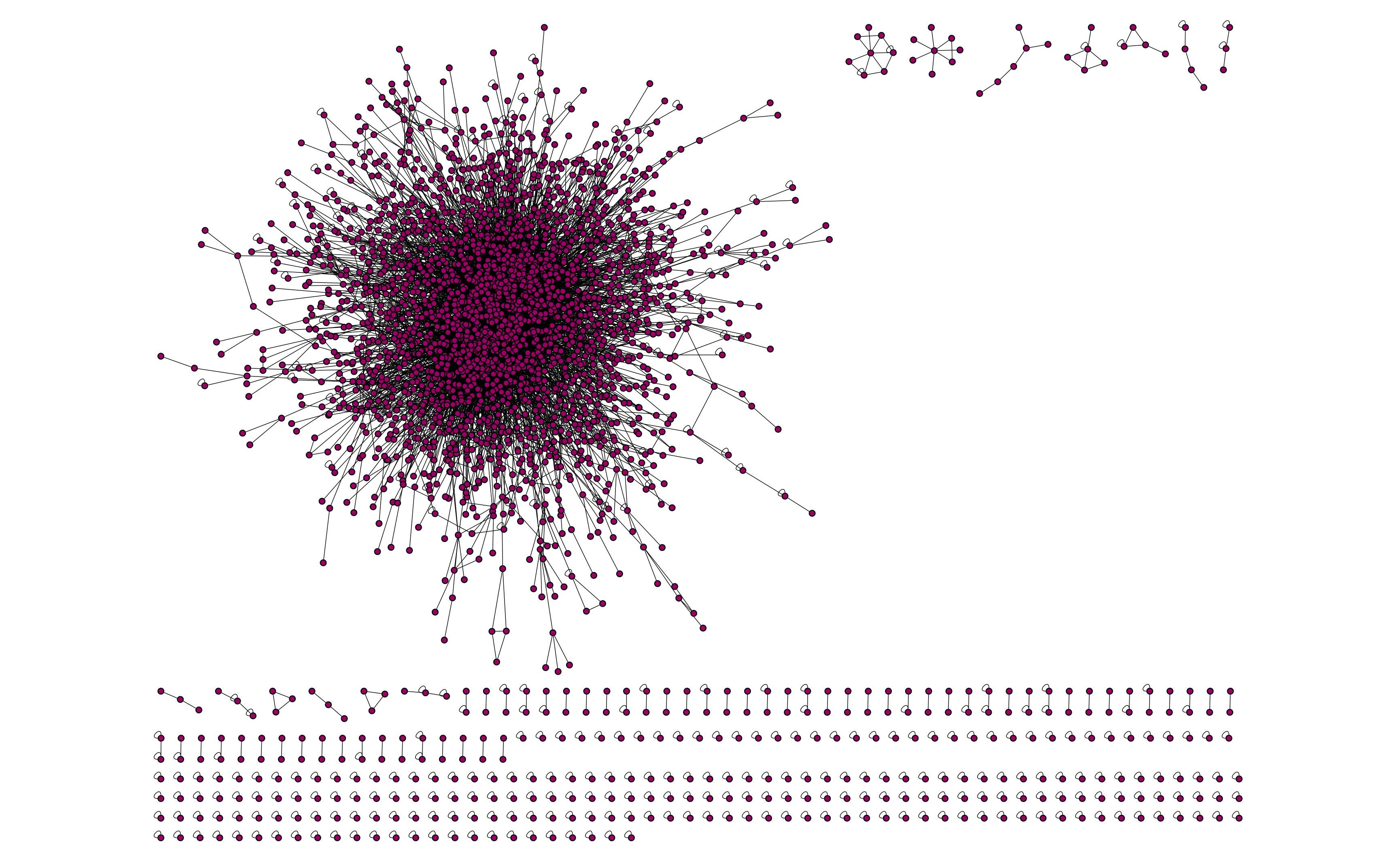
been reported. In the figure below, we illustrate a schematic view of the PPI network which is plotted using Cytoscape
software [2]. The network in question is a fragmented network having a main component with several small
islands. Since, we are going to solve CNDP on this network, the small islands are of no interest and we remove
those islands from the network such that only the main component of the network is remained. Furthermore, there exist
some interactions which are actually self-loops and are removed from the data set. Therefore, the final network of
E. coli consists of 11501 interactions between 2524 proteins. The same progress is done for clearing the PPI network
of S. cerevisiae leading to a final network of 22523 interactions between 5059 proteins.
Essential Proteins
The essential genes information are extracted from DEG database [3]. The data set of E. coli genes essentiality
(being essential or non-essential) includes 11888 experiment reports on 4323 genes. In this data set, there is only
one report for a fraction of genes, more than one experiment has been reported for some of the genes and also
there is no information reported for the remaining genes of E. coli. For genes with more than one report, if
the number of reports about their essentiality/non-essentiality is higher than the number of reports about their
non-essentiality/essentiality, those gene are considered as essentials/non-essentials. If the number of reports on
essentiality and non-essentiality of a gene is equal, we consider the essentiality status of that gene as unknown. In
that way, we have 306 essential genes, 3987 non-essential genes and 30 genes with an unclear status in the data
set extracted from DEG for E. coli. DEG has provided the genes with STRING ids while in DIP the main used
ids are UniProtKB ids. Therefor, to label proteins in the PPI networks as essential or non-essential (and also as
unknown), we use Uniprot ID Mapping [4] to map DIP ids to STRING ids. The status of proteins for which there
is no corresponding gene with reports about its essentiality in DEG, are also considered to be unknown. In this
respect, of 2524 proteins in E. coli PPI network, 371 and 2044 proteins are essential and non-essential, respectively
and there are 109 proteins with the unknown label. The table below, gives an overview of the information about E. coli
and S. cerevisiae PPI networks and the essentiality status of their proteins.
| interactions | proteins | essential proteins | non-essential proteins | unknowns | links | |
|---|---|---|---|---|---|---|
| E. coli | 11501 | 2524 | 371 (15%) | 2044 | 109 | |
| S. cerevisiae | 22523 | 5059 | 967 (19%) | 3500 | 592 |
Source Codes
The source codes for preprocessing the data are as follows.
Processing DIP protein protein interactions:
We first remove any redundancies and self-loops in data of DIP protein protein interactions ( Process_DIP_PPIs.py ).
Then the output file of this step is passed to (Extract_Main_Component.py). In this part of the program we extract only the main component of
the network.
Mapping DIP IDs to STRING IDs
In order to label the proteins in the PPI network as essential (non-essential), we use Uniprot ID Mapping [60] in
the program (Map_DIP_IDs_to_STRING_IDs.py) to map DIP IDs to STRING IDs and then determine the
label of the proteins.
References
[1] Salwinski, L., Miller, C. S., Smith, A. J., Pettit, F. K., Bowie, J. U., and Eisenberg, D. (2004). The database of interacting proteins:
2004 update. Nucleic acids research, 32(suppl-1), D449-D451
[2] Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., ... and Ideker, T. (2003). Cytoscape: a software
environment for integrated models of biomolecular interaction networks. Genome research, 13(11), 2498-2504.
[3] Hao Luo, Yan Lin, Feng Gao, Chun-Ting Zhang and Ren Zhang, (2014) DEG 10, an update of the Database of Essential Genes
that includes both protein-coding genes and non-coding genomic elements. Nucleic Acids Research 42, D574-D580.
[4] Pundir, S., Martin, M. J., O’Donovan, C., and UniProt Consortium. (2016). UniProt tools. Current protocols in bioinformatics,
53(1), 1-29.